DNA BASICS for LAWYERS

 Many of us have seen DNA1 lab reports claiming that a match between our client and a sample of DNA is “5.46 nonillion2 times more probable” than coincidence. We’ve seen charts and data discussing alleles and Y-STR analysis and CODIS hits and lab results from a forensic perspective. This article is not going to discuss the forensic issues involved in defending a DNA case. Rather, this will be a short primer on what DNA itself is, to begin with – a topic that can fill thousands of pages condensed into 1000 words.
Many of us have seen DNA1 lab reports claiming that a match between our client and a sample of DNA is “5.46 nonillion2 times more probable” than coincidence. We’ve seen charts and data discussing alleles and Y-STR analysis and CODIS hits and lab results from a forensic perspective. This article is not going to discuss the forensic issues involved in defending a DNA case. Rather, this will be a short primer on what DNA itself is, to begin with – a topic that can fill thousands of pages condensed into 1000 words.
You may remember back in high school learning about 19th-century monk Gregor Mendel breeding peas that were yellow or green, wrinkly or smooth. Our high school teachers told us that DNA contains genes – a plant might have the gene for yellow peas or the gene for green peas, or it might have both but one of the two is “dominant” and so controls the color. But DNA is so much more than that.
DNA Structure
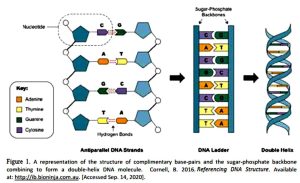
DNA is a very large, highly complex, and infinitely fascinating molecule. Its basic shape is the famous “double helix,” which essentially looks like a twisted ladder. Four molecules known as nucleotide bases, abbreviated A, C, G, and T3, each pair with a specific matching partner – A pairs with T, and C pairs with G. These four bases are the entire alphabet of DNA. Every bit of information encoded by DNA is spelled out by this alphabet of four letters in different combinations. The A-T and C-G base pairs form the rungs of the ladder.
Each nucleotide base is attached to a sugar called deoxyribose and a compound known as a phosphate group. Each of these sugar-phosphate groups “stacks” on its neighbor, creating a long chain known as the sugar-phosphate backbone. Two of these chains make the rails of the ladder, attached to each other by the base-pair rungs. The chains are antiparallel – that is, the sugar-phosphate groups are stacked in opposite directions. Stretched end to end, all the strands of DNA in a single cell about six billion base-pairs – would measure about three meters long4. In order to store all that DNA in a cell only a few millionths as long, the DNA needs to be tightly packed. Proteins known as histones provide this packed structure. The DNA strand wraps itself around histones, forming a structure that is often compared to beads on a string. This structure in turn coils itself again, forming a denser protein mass known as chromatin. When a cell is ready to divide to create two new cells, chromatin will condense further into a familiar structure known as a chromo-some5. Although one of the more well-known terms in genetics, chromosomes are only present during cell division.
 Functions of DNA
Functions of DNA
Physical expression: DNA does, of course, control what a person (or plant, animal, or organism) looks like. Every physical trait you possess is written in your genes. Every gene comes in pairs, one inherited from your mother and one from your father6. Each member of that pair is called an allele, a term you may recognize from your DNA lab reports. Each one of your individual physical traits will be determined by the combination of genes you get from each parent. Take eye color, for example – if your mother and father both have blue eyes and you inherit the blue-eyed allele from both, you will have blue eyes. But if your father has brown eyes, you may end up with a pair of mismatched alleles. In that case, one of the alleles may be dominant and override the other7. Observable physical characteristics expressed by genes are known as the phenotype. The genes themselves are known as the genotype.
Biological function and development: The alphabet of DNA can be broken down into three-letter “words” known as codons. Each codon is exactly three base pairs long, and each combination of three letters specifies the creation of one of 20 specific amino acids. These amino acids, in turn, combine in various combinations to form the hundreds of thousands of proteins the human body requires. These proteins drive the countless biological functions the body constantly performs or they are used to build or repair the body itself.

Gene regulation: Proteins created by DNA can be used to “switch” other genes on or off. In a sense, this is DNA acting as its own metadata.
Self-replication and repair: In order for cells to divide, for an organism to develop, or for a person to grow, DNA must be able to copy itself for every newly-created cell. DNA can even repair itself. Because a length of DNA is made of up two complementary strands, an undamaged strand can be used as a template to repair a damaged strand. Each individual cell suffers and repairs DNA damage tens of thousands of times every day, making DNA an incredibly robust method of information storage.
DNA Analysis
When our clients submit a DNA sample, whether by blood, cheek swab, saliva, or another method, the cells collected need to be broken down and the DNA inside extracted. The amount available from the few cells in the sample is too small to analyze. That small amount must be repeatedly copied through a process known as the Polymerase Chain Reaction or PCR8 to make enough DNA to test. The PCR process, in essence, separates the DNA ladder into two separate single strands. Those two single strands of DNA are each copied, yielding two full strands. This process is repeated, two full strands yielding four, then four strands yielding eight, and so on.
The actual comparison is done using short tandem repeats (STR). STRs are sequences of several base pairs that repeat between a few and a few dozen times. Every individual has a unique “fingerprint” comprised of a different number of repeats at specific locations (loci) in the genome. The more of these loci examined, the more statistically accurate the test. This process is sometimes called Y-STR when the analysis is restricted to the Y (male-only) chromosome. The trouble for our clients often begins when their DNA is compared with a sample from the crime scene or victim, or with the FBI’s CODIS database. Figure 4 is an example of an STR profile known as an electropherogram. Examining just the top half for brevity, we see different loci named D19S433, vWA, TPOX, D18S51. The length of each bit of DNA is given by the scale on the Y axis, with 0 base pairs (bp) and 1700 bp noted on the scale. For this example, we would say that the subject’s genotype is 13, 14 at locus D19S433; 14, 17 at locus vWA; 11, 11 at TPOX; 13, 16 at D18S51; etc. A locus with two numbers (e.g. 13, 14) indicates that the source inherited one of each allele from each parent. A locus with one number (e.g. 11, 11) shows that the individual inherited the same allele from each parent.
The trouble for our clients often begins when their DNA is compared with a sample from the crime scene or victim, or with the FBI’s CODIS database. Figure 4 is an example of an STR profile known as an electropherogram. Examining just the top half for brevity, we see different loci named D19S433, vWA, TPOX, D18S51. The length of each bit of DNA is given by the scale on the Y axis, with 0 base pairs (bp) and 1700 bp noted on the scale. For this example, we would say that the subject’s genotype is 13, 14 at locus D19S433; 14, 17 at locus vWA; 11, 11 at TPOX; 13, 16 at D18S51; etc. A locus with two numbers (e.g. 13, 14) indicates that the source inherited one of each allele from each parent. A locus with one number (e.g. 11, 11) shows that the individual inherited the same allele from each parent.
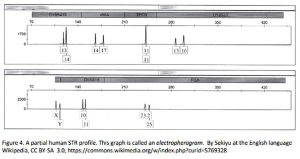
This genotype is then compared with the target sample. Because the frequency that each allele appears in a general population is also known, every matching locus increases the statistical confidence that the subject and target are the same people. Using the population frequency of each allele, the tester can calculate the chance that each of those alleles will all appear together in a random member of the population. This number is known as random match possibility or RMP. In the example used at the beginning of this article, 24 separate loci were compared, yielding the result that a match, in that case, is 5.46 nonillion times more probable than coincidence.
Of course, these results get vastly more complicated. In many cases a DNA sample may have a mix of multiple contributors, making both physical and statistical analysis much more complicated. Sometimes, due to degraded or insufficient samples, an allele may be missing from the data leaving only a partial profile. The graphs themselves may be corrupted or obscured by artifacts, background noise, defects in either equipment or procedures, air bubbles, extremely small (or too much!) DNA sample amounts, simple interpretation bias, and more. On top of that, the Government loves to make hay about the giant numbers 9, 10, and try to claim that the tests say more than they actually do. DNA evidence, while compelling and probably one of the most accurate and reliable of the forensic tests, is still not infallible. As with any forensic result – when in doubt, get an expert!
1. Deoxyribonucleic acid.
2. That’s 30 zeros, or in this example from an actual case, 1 in 5,460,000,000,000,000,000,000,000,000 ,000. For comparison, there have only been about 400 or so quadrillion (400,000,000,000,000,000) seconds since the birth of the universe. You have a 1 in 292,000,000 chance of winning the Powerball jackpot.
3. Adenosine, Cytosine, Guanine and Thymine.
4. To impart a sense of just how efficiently DNA is packed into the cell: all the DNA in your body stretched out would be 10 billion miles long. That’s a bit more than the distance from Earth to Pluto and back.
5. Humans have 23 pairs of chromosomes, for a total of 46.
6. With some exceptions such as mitochondrial DNA (mtDNA), which are beyond the small scope of this article.
7. In the case of eyes, the brown-eyed gene is dominant so a person with one blue and one brown allele will have brown eyes. Some alleles are semi-dominant and result in a mixed physical trait. For example, a plant with red flowers and a plant with white flowers may breed a plant with pink flowers.
8. PCR was invented by biochemist and LSD aficionado Kary Mullis in 1983, for which he won the Nobel Prize.
9. The random match possibility is actually the probability that the suspect has the same DNA genotype and is not the source of the sample. Beware the transposition fallacy, aka “prosecutor’s fallacy”: a 1 in 5.46 nonillion chance that the suspect would match if he is not the source is not always the same as saying 5.46 nonillion-to-1 that the suspect is the source.
10. There is also a “defendant’s fallacy,” which goes something like: with a random match possibility of one in a million, there could be three unrelated matches in a city with a population of three million, therefore there is a two in three chance the defendant is not the source.
The Fortress Law Office
4124 Erie St
Willoughby, OH 44094
(440) 340-1740
https://www.bangerterlaw.com/


